© Hong Kong Academy of Medicine. CC BY-NC-ND 4.0
ORIGINAL ARTICLE
Prevalences of levofloxacin resistance and pncA
mutation in isoniazid-resistant Mycobacterium tuberculosis in Hong Kong
Kevin KM Ng, MB, ChB, FRCPath; Peter CW Yip, PhD; Patricia KL Leung, MPhil
Department of Public Health Laboratory Services Branch, Centre for Health Protection, Department of Health, Hong Kong
Corresponding author: Dr Kevin KM Ng (kevinkmng@yahoo.com.hk)
Abstract
Introduction: In 2018, the World Health
Organization recommended a 6-month treatment
regimen that included levofloxacin and pyrazinamide
for isoniazid-resistant Mycobacterium tuberculosis
without rifampicin resistance (Hr-TB). Susceptibility
testing for both drugs is not routinely performed
for Hr-TB in Hong Kong. This study examined
the prevalences of levofloxacin and pyrazinamide
resistances in Hr-TB and explored associated risk
factors.
Methods: All Hr-TB isolates archived during 2018
were retrieved. Isolates were de-duplicated to
identify unique cases. Levofloxacin susceptibility
testing was performed using the MGIT 960 System;
pncA gene sequencing was used as a surrogate
indicator of pyrazinamide susceptibility. Previous
laboratory records for each case were analysed.
Results: In total, 160 phenotypic Hr-TB cases were
identified from among 3411 patients with tuberculosis
(4.7%). Among these, 157 were analysed, revealing
0.6% (n=1) levofloxacin resistance and 4.5% (n=7) pyrazinamide resistance, respectively. Independent
risk factors associated with pncA mutations included
history of tuberculosis in the affected patient and
isoniazid poly-resistance (ie, double and triple
resistances), but not mono-resistance.
Conclusion: For Hr-TB in Hong Kong, levofloxacin
resistance is rare and pyrazinamide resistance-associated
pncA mutations are uncommon. Routine
susceptibility testing for these drugs is not indicated
unless related risk factors are identified.
New knowledge added by this study
- Levofloxacin (LVX) resistance is rare (0.6%) and pyrazinamide (PZA) resistance-associated pncA mutations are uncommon (4.5%) among isoniazid-resistant, rifampicin-susceptible Mycobacterium tuberculosis (Hr-TB) isolates in Hong Kong.
- Risk factors for pncA mutations in Hr-TB include history of tuberculosis in the affected patient and isoniazid poly-resistance.
- Clinicians could initiate empirical treatment for patients with Hr-TB without routine susceptibility testing for LVX and PZA.
- Susceptibility testing for LVX and PZA could be considered in patients with Hr-TB and additional risk factors; or when clinical, radiological, and microbiological responses are suboptimal during early follow-up.
Introduction
There has been a gradual decline in the global
tuberculosis (TB) incidence and mortality rates
over time, by approximately 2% and 3% per year,
respectively.1 Despite this trend, TB remains a leading
cause of death, and Mycobacterium tuberculosis drug
resistance continues to be a major public health issue.
Globally, isoniazid (INH)-resistant, rifampicin (RIF)-susceptible TB (Hr-TB) is the most common drug-resistant
form of disease.2 In 2017, the rates of Hr-TB were 7.1% among patients with new TB diagnoses
and 7.9% in patients who had previously undergone treatment for TB.1 In Hong Kong, the respective
rates were 5.3% and 9.5%, with a combined rate of
5.7% in 2016.3 The Hr-TB is associated with worse
clinical outcome and development of multidrug
resistance (MDR),4 5 although the findings have been
inconsistent.6 7
Isoniazid constitutes a key component in
the treatment of drug-susceptible TB, through
its inhibition of mycolic acid biosynthesis in the
mycobacterial cell wall. Over 85% of INH resistance
is conferred by mutations residing in the katG gene
and inhA promoter region,8 leading to high and low levels of resistance, respectively. These mutations
can readily be detected by commercial molecular
line-probe assays. The level of resistance can also
vary according to the co-occurrences of less common
mutations in other regions.
For the treatment of Hr-TB, current World
Health Organization (WHO) guidelines recommend
6 months of RIF, pyrazinamide (PZA), ethambutol,
and levofloxacin (LVX). The guidelines also
recommend examination of resistances towards
fluoroquinolones and PZA, prior to treatment.9
In our locality, routine testing of LVX and
PZA susceptibility has not been implemented for
the treatment of Hr-TB. To determine the most
cost-effective testing strategy, this study reviewed
an annual collection of Hr-TB isolates, with the aim
of determining the prevalences of LVX resistance
and pncA mutations (as a molecular marker of
PZA resistance in Hr-TB) and identifying factors
associated with these resistances.
Methods
Case identification and data collection
Data were reviewed from the TB Reference
Laboratory of the Department of Health. This
laboratory processes local M tuberculosis strains
from specimens collected in out-patient clinics
and in-patient hospitals in both public and private
sectors. All phenotypic Hr-TB isolates in 2018 were
identified and de-duplicated using unique patient
identifiers. The following data were included in this
analysis: basic patient demographics, phenotypic susceptibility results of first-line drugs (INH at
critical concentrations 0.1 μg/mL and 0.4 μg/mL as
recommended by the WHO, RIF, streptomycin, and
ethambutol), and genotypic susceptibility results
(inhA promoter region, katG codon 315, and rpoB
hotspot region) from the GenoType MTBDRplus
assay, version 2.0 (Hain Lifescience). Any discrepant
or unsuccessful test results were resolved by the agar
proportion method and/or DNA sequencing; when
applicable, these additional data were reported as the
final results. All available data from the Laboratory
Information System were reviewed for each patient
to determine any history of TB, fluorescence
microscopy results (according to Global Laboratory
Initiative grading10), site of isolation (pulmonary
versus extrapulmonary), any documented sputum
culture conversion and the duration (ie, time between
the date of first positive culture for current infection
and the date of consistently negative culture), and
microbiological outcome.
Levofloxacin and pyrazinamide susceptibility
testing and pncA gene sequencing
Selected M tuberculosis strains were retrieved
from the laboratory archive and sub-cultured,
then subjected to LVX susceptibility testing
using the BD BACTEC MGIT 960 system
(Becton, Dickson and Company), in accordance
with the manufacturer’s instructions. The LVX
critical concentration at 1.0 μg/mL was used as
recommended by the WHO. DNA extraction was
performed using GenoLyse (Hain Lifescience).
The pncA gene was amplified with the primers
pncA-1F (5′-CGCTCAGCTGGTCATGTTC-3′) and
pncA-1R (5′-CCCACCGGGTCTTCGAC-3′) to
produce an amplicon of 798 bp, using the GeneAmp
PCR System 9700 (Applied Biosystems). Each 25-μL
reaction mixture contained 12.5 μL of GoTaq G2 Hot
Start Colorless Master Mix (Promega) [1×], 0.25 μL
of each primer (1.0 pM/μL), 9.5 μL PCR-grade water,
and 2.5 μL of template DNA. Reaction mixtures
were amplified using the following protocol:
2 minutes at 95°C; 35 cycles of 1 min at 95°C,
1 minute at 65°C, and 1 minute at 72°C; and 10 minutes
at 72°C. Sequencing was performed using the
3730 × l DNA Analyzer (Applied Biosystems) with the
same primers. Resulting sequences were compared
with the sequence of wild-type M tuberculosis
H37Rv for detection of pncA mutations. Strains
with detected pncA mutations were subjected to
further analysis of PZA susceptibility via the MGIT
960 system in accordance with the manufacturer’s
instructions, with a critical concentration of
100 μg/mL.
Statistical analysis
Univariate analysis comprised odds ratio calculations
and Fisher’s exact test. Firth logistic regression was employed for multivariable analysis because of
the low outcome frequency. IBM SPSS Statistics
Subscription (Windows version, IBM Corp, Armonk
[NY], United States) was used for data analysis. A P
value of <0.05 was considered statistically significant.
This article complies with the STROBE Statement
reporting guidelines.
Results
In total, 8865 M tuberculosis isolates from 3411
patients were processed in 2018. Susceptibility
profiles were available for all except repeated strains
collected from the same patient within 3 months.
De-duplication of 393 isolates yielded 160 patients
with phenotypic Hr-TB, amounting to a case rate
of 4.7%. rpoB hotspot mutations were identified
in five cases by GenoType MDRTBplus, three of
which were confirmed to confer RIF resistance
according to rpoB sequencing results. These three
cases were considered to be RIF-resistant and
excluded from further analysis. The mean age of the
patients in the remaining 157 cases was 61.3 years
(range, 15-95 years); the male to female ratio was
1.8. Pulmonary TB was present in 90.4% of affected
patients (n=142), while 9.6% of affected patients
(n=15) had extrapulmonary involvement. There was
a documented history of TB by positive culture in
4.5% of affected patients (n=7). Microscopy results
were available for cases in which direct specimens
were received by our laboratory at the time of initial
diagnosis (n=45). The majority of specimens was
acid-fast bacillus smear-negative (n=33), followed by
1-4 acid-fast bacilli per length (n=4), scanty (n=4),
1+ (n=2), and 2+ and 3+ (n=1 each). In terms of
microbiological outcome, 76.4% of affected patients
(n=120) had documented sputum culture conversion
without recurrence after follow-up of at least
9 months, among which 106 attained conversion
within 5 months after diagnosis. The median interval
required for culture conversion was 72.5 days (range,
9-622 days). No clearance was documented for
patients in the remaining 37 cases, for whom no
follow-up specimens were received for >1 year.
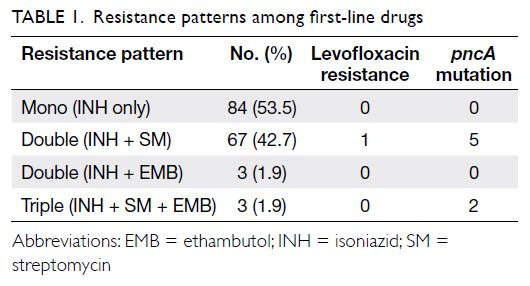
Table 1 shows the patterns of resistance among
first-line drugs and distributions of cases with LVX
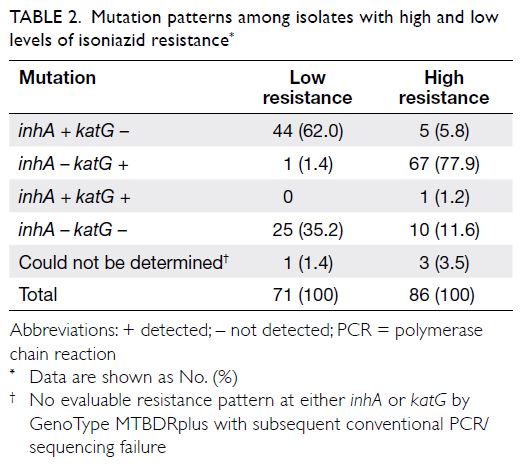
resistance and pncA mutations. Table 2 shows the
patterns of mutations detected. With respect to
INH, 45.2% (n=71) of the isolates exhibited low-level
resistance (minimum inhibitory concentration
0.1-0.4 μg/mL) and 54.8% (n=86) of the isolates
exhibited high-level resistance (minimum inhibitory
concentration >0.4 μg/mL).
For LVX, only one strain was resistant, while
155 were sensitive (one isolate failed to be recovered).
The number of resistant cases was insufficient for
correlation analysis. pncA mutations were detected
in 4.5% (7/157 cases), and all detected mutations were
previously identified as PZA resistance–related. All isolates with pncA mutations were phenotypically
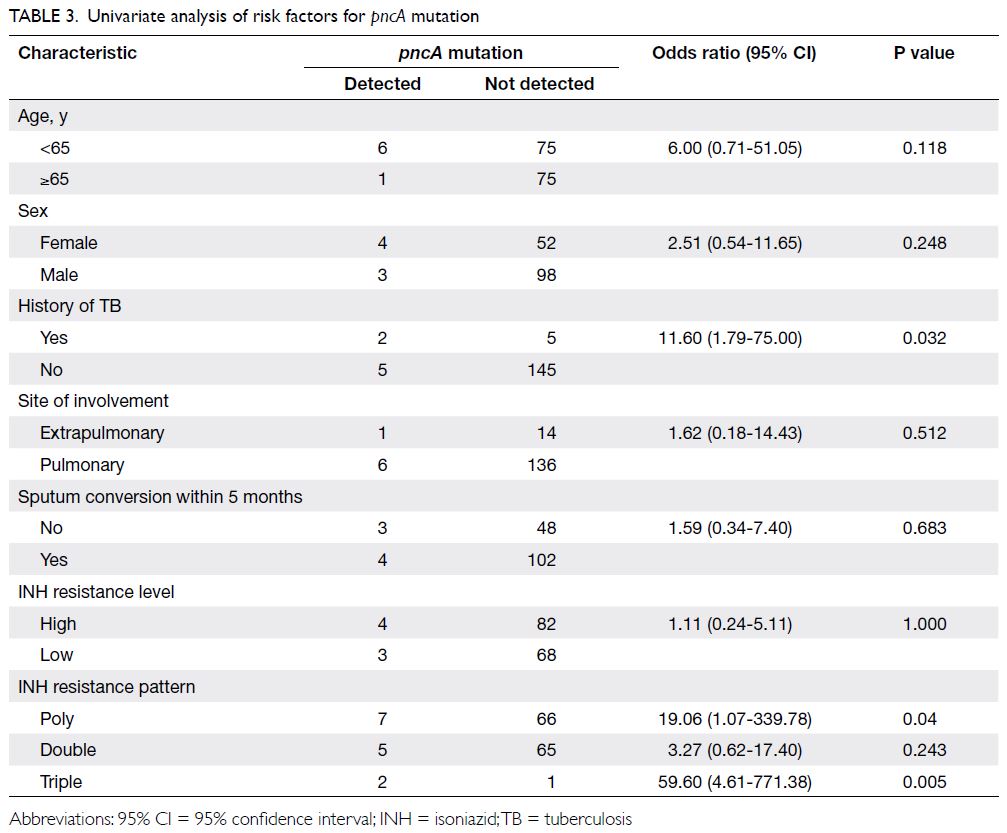
resistant to PZA. Univariate analysis showed that
pncA mutations were associated with documented
history of TB in the affected patient (odds ratio
[OR]=11.60, 95% confidence interval [CI]=1.79-75.00; P=0.03) and INH poly-resistance (OR=19.06,
95% CI=1.07-339.78; P=0.04) but not mono-resistance,
as shown in Table 3. In multivariable
logistic regression, pncA mutations were associated
with documented history of TB in the affected
patient (OR=18.03, 95% CI=2.25-153.85; P=0.008),
INH triple resistance (OR=409.11, 95% CI=6.52
to >1000; P<0.001), and INH double resistance
(OR=13.50, 95% CI=1.43 to >1000; P=0.019).
Discussion
Levofloxacin resistance
In this study, the frequency of LVX resistance was
very low in Hr-TB. Levofloxacin is an important drug
in the management of drug-resistant TB. In a 2009
study in Shanghai, Xu et al11 estimated the rates of
LVX resistance to be 1.9% in strains pan-susceptible to
first-line drugs, 6.7% in INH mono-resistant strains,
and ≤25% in MDR-TB. Independent risk factors associated with LVX resistance included MDR, RIF
mono-resistance, poly-resistance (resistance to ≥2
first-line drugs, but not MDR), age ≥46 years, and TB
re-treatment, but not INH mono-resistance. These
findings were consistent with the results of a 2017
study in Ningbo (a city near Shanghai), whereby Che
et al12 revealed no LVX resistance in strains pan-susceptible
to first-line drugs, 3% in strains with any
resistance to first-line drugs except MDR, and ≤30%
in MDR-TB. Most fluoroquinolone resistance was
present in the MDR group. Che et al12 also found that
prevalence of LVX resistance in MDR-TB increased
with duration of inappropriate treatment before LVX;
this relationship was stronger than any relationships
with previous fluoroquinolone exposures. Further
studies are needed to determine whether this finding
also applies to Hr-TB. Levofloxacin resistance is
uncommon in non-MDR strains, especially in
the present study. Since 2015, our laboratory has
performed LVX susceptibility testing on 640 Hr-TB
strains, identifying only two resistant cases (0.31%;
unpublished data), including the one in this study.
Clinicians could treat affected patients empirically, in accordance with WHO recommendations;
further testing could be arranged based on clinical,
radiological, and microbiological responses during
early follow-up. In patients with Hr-TB that exhibits
LVX resistance or poly-resistance to other primary
agents, individualised adjustment of the treatment
regimen is recommended in accordance with WHO
guidelines9 (eg, exclusion of LVX or inclusion of
second-line TB medicines). In our single case
with LVX resistance, the patient had no history
of TB. His sputum acid-fast bacillus smear result
was 2+. The Hr-TB strain isolated was initially
LVX-sensitive without mutations in rpoB, gyrA, or
gyrB on diagnosis and follow-up at 8 months.
The patient continued to exhibit sputum culture-positive
results after 15 months of treatment; further
analysis revealed that the isolate had acquired
gyrA mutation (detected by line-probe assays) and
LVX phenotypic resistance. It then developed into
an MDR strain; the patient eventually achieved
culture conversion at approximately 18 months
after diagnosis. Underlying hetero-resistance was a
possible contributing factor.
Pyrazinamide resistance
Phenotypic PZA susceptibility testing has often been
problematic. pncA mutations constitute the most
common and primary determinant of PZA resistance,
with reported sensitivity of approximately 80% to
95% and specificity of approximately 85% to 100%.13 14
Resistance-conferring mutations are known to be
diverse and scattered over the gene’s full length. Our
seven isolates all had unique mutations (Gln10Arg,
His51Tyr, Trp68Stop, Thr47Pro, Ala102Thr,
Asp63Gly, and Val139Ala), which were associated
with PZA resistance at a high probability of 0.985
according to available literature15 (except Val139Ala,
probability of resistance=0.783). These seven isolates
were also confirmed to be phenotypically resistant to
PZA in our study.
Thus far, investigations of phenotypic and
genotypic PZA resistance have generally focused
on MDR-TB. Concerning non-MDR-resistant
strains, there is considerable geographical variation
in the reported rates, from 2.92% to 4.8% in the
United States,16 17 to 24% in the Western Pacific and
75% in South East Asia.18 For Hr-TB in particular,
phenotypic PZA resistance was reported in 2% of
INH mono-resistant strains in South Africa,19 no
PZA resistance was detected (phenotypic or pncA
mutation) in Georgia (Eastern Europe) among
Hr-TB strains,20 and ≤14.9% of Hr-TB strains
exhibited pncA mutations in Vietnam.21 In our
study, we observed that 4.5% of Hr-TB isolates had
pncA mutations. Such variation may be related to
multiple factors, including differences in inclusion
criteria, testing method, treatment, infection control
practice, and disease burden. In terms of risk factors,
we found that history of TB in the affected patient
and poly-resistance (but not mono-resistance) were
significantly associated with pncA mutations. These
results are consistent with findings from previous
studies, which also showed an association with
MDR.21 22 23 Clinicians are advised to consider the
possibility of PZA resistance in patients with Hr-TB
and these risk factors. Routine testing for LVX or
PZA susceptibility in patients with INH mono-resistant
TB alone (ie, no other risk factors) is not
considered cost-effective, based on our findings.
Outcomes and treatment of isoniazidresistant
Mycobacterium tuberculosis without rifampicin resistance
In our study, most patients with Hr-TB had a
satisfactory microbiological outcome without
recurrence. However, the outcome was unknown in
23.6% of cases. Most of these involved patients were
diagnosed and managed in hospitals, where only
positive isolates were subjected to reference testing
at our laboratory. Based on the recommended
management regimen for TB in Hong Kong, these patients would have been followed up until sputum
culture conversion. Considering the time elapsed
since the latest positive laboratory result, the patients
in most cases with unknown outcomes likely already
had culture conversion.
In general, Hr-TB is presumably associated
with higher rates of treatment failure and MDR
acquisition, compared with drug-susceptible
strains.4 5 Important considerations for disease
control include recognition of risk factors
associated with Hr-TB (eg, history of TB treatment,
incarceration in prisons, and homelessness7 24),
early detection of INH resistance, exclusion of RIF
resistance by appropriate molecular methods (eg,
line-probe assays),2 and compliance with current
treatment guidelines. With the maturation of whole-genome
sequencing, the technology is becoming
integrated into the local routine drug susceptibility
testing protocol for all M tuberculosis isolates;
multiple and uncommon molecular markers of drug
resistance might thus be identified earlier in a single
set of assays for proper treatment guidance.
Strengths and limitations
Because our laboratory serves as a local TB reference
laboratory, this study was able to use a representative
M tuberculosis collection that reflected the status
of Hr-TB in Hong Kong. Furthermore, because
our laboratory serves as a supranational reference
laboratory, we were able to perform an array of
confirmatory tests in accordance with WHO
recommendations, thus ensuring reliable results.
However, this study had some limitations. First,
it lacked data regarding microscopy results, drug
treatment, and clinical outcome, thus preventing
a more comprehensive assessment. Second, the
small sample size and low outcome frequencies of
LVX and PZA non-susceptibilities led to limited
correlation analysis. Third, less common mutations
mediating INH resistance were not examined to
determine their prevalences; Hr-TB cases with these
mutations would only be identified on the basis of
phenotypic results. A substantial proportion of cases
(22.3%; n=35) had negative GenoType MDRTBplus
results. Because this test only detects the most common mutations at the inhA promoter region
and katG codon 315, other relevant mutations might
have been missed. Furthermore, in 3.2% (n=5) of
cases with high-level INH resistance, mutations
were detected in the inhA promoter region alone.
The availability of preliminary information before
receiving the INH phenotypic susceptibility results
might lead to mismanagement of patient treatment
(eg, INH inclusion in the therapeutic regimen).
Fourth, low-level RIF resistance mediated by
mutations outside the rpoB hotspot region and
undetected by phenotypic testing was not ruled out in this study. Inclusion of such isolates might have
led to overestimation of true resistance in Hr-TB.
Fifth, PZA resistance could be mediated by other
less common mutations, which were not examined
by performing phenotypic PZA susceptibility
testing on all Hr-TB isolates; thus, we may have
underestimated its prevalence. Our laboratory has
found that all PZA-resistant M tuberculosis isolates
thus far could be confirmed through pncA mutation
analysis. Nevertheless, we presume that these
limitations do not greatly affect the reliability and
overall interpretation of our findings.
Conclusion
In Hong Kong, the rate of Hr-TB was 4.7% among
active TB cases in 2018. Levofloxacin and PZA
resistances among Hr-TB were uncommon (0.6%
and 4.5%, respectively). History of TB in the affected
patient and INH poly-resistance, including both
double and triple resistances, were independent risk
factors for pncA mutations. The findings from this
study do not support routine susceptibility testing
for these two drugs in patients with INH mono-resistant
TB who lack additional risk factors.
Author contributions
Concept or design: KKM Ng.
Acquisition of data: All authors.
Analysis or interpretation of data: All authors.
Drafting of the manuscript: KKM Ng.
Critical revision of the manuscript for important intellectual content: KKM Ng.
Acquisition of data: All authors.
Analysis or interpretation of data: All authors.
Drafting of the manuscript: KKM Ng.
Critical revision of the manuscript for important intellectual content: KKM Ng.
All authors had full access to the data, contributed to the study, approved the final version for publication, and take
responsibility for its accuracy and integrity.
Conflicts of interest
All authors have disclosed no conflicts of interest.
Acknowledgement
The authors acknowledge the excellent work and contribution by staff, especially Mr Steven CW Lui and Ms Angela WL
Lau, at the Tuberculosis Laboratory and Special Investigation
Laboratory of Public Health Laboratory Services Branch,
Centre for Health Protection, Department of Health, Hong
Kong SAR Government.
Funding/support
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Ethics approval
Ethics approval for this study was obtained from the Ethics Committee of the Department of Health, Hong Kong SAR
Government (Ref: LM 425/2019). All included patients
consented to testing for tuberculosis.
References
1. World Health Organization. Global tuberculosis
report 2018. Available from: https://www.who.int/tb/publications/global_report/en/. Accessed 19 Sep 2019.
2. Romanowski K, Campbell JR, Oxlade O, Fregonese F, Menzies D, Johnston JC. The impact of improved
detection and treatment of isoniazid resistant tuberculosis
on prevalence of multi-drug resistant tuberculosis: a
modelling study. PLoS One 2019;14:e0211355. Crossref
3. Tuberculosis & Chest Service of Department of Health,
Hong Kong SAR Government. Annual report 2016. Available from: https://www.info.gov.hk/tb_chest/doc/Annual_Report_2016.pdf. Accessed 19 Sep 2019.
4. Gegia M, Winters N, Benedetti A, van Soolingen D,
Menzies D. Treatment of isoniazid-resistant tuberculosis
with first-line drugs: a systematic review and meta-analysis.
Lancet Infect Dis 2017;17:223-34. Crossref
5. Karo B, Kohlenberg A, Hollo V, et al. Isoniazid (INH)
mono-resistance and tuberculosis (TB) treatment success:
analysis of European surveillance data, 2002 to 2014. Euro
Surveill 2019;24:1800392. Crossref
6. Salindri AD, Sales RF, DiMiceli L, Schechter MC, Kempker RR, Magee MJ. Isoniazid monoresistance and
rate of culture conversion among patients in the State of
Georgia with confirmed tuberculosis, 2009-2014. Ann Am
Thorac Soc 2018;15:331-40. Crossref
7. Cattamanchi A, Dantes RB, Metcalfe JZ, et al. Clinical
characteristics and treatment outcomes of isoniazid-monoresistant
tuberculosis. Clin Infect Dis 2009;48:179-85. Crossref
8. Seifert M, Catanzaro D, Catanzaro A, Rodwell TC.
Genetic mutations associated with isoniazid resistance
in Mycobacterium tuberculosis: a systematic review. PLoS
One 2015;10:e0119628. Crossref
9. World Health Organization. WHO consolidated guidelines on drug-resistant tuberculosis treatment. Available from:
https://www.who.int/tb/publications/2019/consolidated-guidelines-drug-resistant-TB-treatment/en/. Accessed 19
Sep 2019. Crossref
10. Global Laboratory Initiative. Mycobacteriology Laboratory Manual. 1st ed. Global Laboratory Initiative; 2014
11. Xu P, Li X, Zhao M, et al. Prevalence of fluoroquinolone resistance among tuberculosis patients in Shanghai, China.
Antimicrob Agents Chemother 2009;53:3170-2. Crossref
12. Che Y, Song Q, Yang T, Ping G, Yu M. Fluoroquinolone
resistance in multidrug-resistant Mycobacterium
tuberculosis independent of fluoroquinolone use. Eur
Respir J 2017;50:1701633. Crossref
13. Streicher EM, Maharaj K, York T, et al. Rapid sequencing of the Mycobacterium tuberculosis pncA gene for detection of pyrazinamide susceptibility. J Clin Microbiol 2014;52:4056-7. Crossref
14. Khan MT, Malik SI, Ali S, et al. Pyrazinamide resistance
and mutations in pncA among isolates of Mycobacterium
tuberculosis from Khyber Pakhtunkhwa, Pakistan. BMC
Infect Dis 2019;19:116. Crossref
15. Miotto P, Cabibbe AM, Feuerriegel S, et al. Mycobacterium tuberculosis pyrazinamide resistance determinants: a
multicenter study. mBio 2014;5:e01819-4.Crossref
16. Kurbatova EV, Cavanaugh JS, Dalton T, Click ES, Cegielski
JP. Epidemiology of pyrazinamide-resistant tuberculosis in
the United States, 1999-2009. Clin Infect Dis 2013;57:1081-93. Crossref
17. Budzik JM, Jarlsberg LG, Higashi J, et al. Pyrazinamide resistance, Mycobacterium tuberculosis lineage and
treatment outcomes in San Francisco, California. PLoS
One 2014;9:e95645. Crossref
18. Whitfield MG, Soeters HM, Warren RM, et al. A global perspective on pyrazinamide resistance: systematic review
and meta-analysis. PLoS One 2015;10:e0133869. Crossref
19. Whitfield MG, Streicher EM, Dolby T, et al. Prevalence of pyrazinamide resistance across the spectrum of drug
resistant phenotypes of Mycobacterium tuberculosis.
Tuberculosis (Edinb) 2016;99:128-30. Crossref
20. Sengstake S, Bergval IL, Schuitema AR, et al. Pyrazinamide resistance-conferring mutations in pncA and the
transmission of multidrug resistant TB in Georgia. BMC
Infect Dis 2017;17:491. Crossref
21. Huy NQ, Lucie C, Hoa T, et al. Molecular analysis of
pyrazinamide resistance in Mycobacterium tuberculosis
in Vietnam highlights the rate of pyrazinamide resistance-associated
mutations in clinical isolates. Emerg Microbes
Infect 2017;6:e86. Crossref
22. Li D, Hu Y, Werngren J, et al. Multicenter study of the emergence and genetic characteristics of pyrazinamide-resistant
tuberculosis in China. Antimicrob Agents
Chemother 2016;60:5159-66. Crossref
23. Chiu YC, Huang SF, Yu KW, Lee YC, Feng JY, Su JY. Characteristics of pncA mutations in multidrug-resistant
tuberculosis in Taiwan. BMC Infect Dis 2011;11:240. Crossref
24. Smith CM, Trienekens SC, Anderson C, et al. Twenty years and counting: epidemiology of an outbreak of isoniazid-resistant
tuberculosis in England and Wales, 1995 to 2014.
Euro Surveill 2017;22:30467. Crossref