Hong Kong Med J 2014;20:251–4 | Number 3, June 2014
DOI: 10.12809/hkmj134087
© Hong Kong Academy of Medicine. CC BY-NC-ND 4.0
CASE REPORT
Live birth following double-factor
pre-implantation genetic diagnosis for both reciprocal
translocation and alpha-thalassaemia
Vivian CY Lee, FHKAM (Obstetrics and
Gynaecology); Judy FC Chow, MPhil; Estella YL Lau, PhD; William SB
Yeung, PhD; Ernest HY Ng; MD
Department of Obstetrics and Gynaecology,
The University of Hong Kong, Queen Mary Hospital, Pokfulam, Hong
Kong
Corresponding author: Dr Vivian CY Lee (v200lee@hku.hk)
Abstract
We report a live birth from a couple with
two genetic diseases, namely: reciprocal translocation carrier
and alpha-thalassaemia trait, following pre-implantation genetic
diagnostic tests. This is the first case in Hong Kong in which
the technique of using one blastomere biopsy for two diseases
was established, using array comparative genomic hybridisation
and polymerase chain reaction.
Introduction
In this report, we present a couple who
requested a double-factor pre-implantation genetic diagnosis (PGD)
for both reciprocal translocation and alpha-thalassaemia.
Case report
Our patient, aged 36 years, enjoyed good
past health and attended the subfertility clinic for recurrent
miscarriage (5 times within 8 years). She had four spontaneous
conceptions between 1997 and 2004 but all ended as first-trimester
miscarriages. After 2004, she suffered from secondary subfertility
and conceived again in 2007 following ovarian stimulation and
intrauterine insemination. The fifth pregnancy again ended with
first-trimester miscarriage.
She was subsequently referred to a clinical
geneticist and found to be a carrier of a balanced reciprocal
translocation 46,XX,t(2;10)(q33;q21.2). The husband had normal
karyotypes and other relevant investigations for recurrent
miscarriage were all negative. Both partners were
alpha-thalassaemia trait carriers (South East Asia [SEA] type) as
the genotype report revealed heterozygous alpha SEA type deletion.
They were therefore referred to us for PGD.
Baseline investigations showed early
follicular follicle-stimulating hormone levels of 6.5 IU/L and an
antral follicle count of 19. The couple was counselled about the
procedure and risks of PGD. It was decided to biopsy two
blastomeres, so as to perform polymerase chain reaction (PCR) for
alpha-thalassaemia on one of them, and carry out fluorescent
in-situ hybridisation (FISH) for the reciprocal translocation on
the other.
The first cycle of in-vitro fertilisation
(IVF) and intra-cytoplasmic sperm injection (ICSI) was carried out
in November 2010. After 10 days of ovarian stimulation, 12 oocytes
were retrieved and 11 were in metaphase II for ICSI. Ten were
normally fertilised and seven day-3 embryos were available for
embryo biopsy. Two embryos were subsequently shown to be normal
for FISH signals and either normal or heterozygous for
alpha-thalassaemia SEA deletion. On day 5, there was only one
fair-quality embryo at the morula stage for transfer, but the
patient failed to conceive in that cycle.
She underwent a second IVF/ICSI/PGD cycle
in January 2012. After 11 days of ovarian stimulation, 13 oocytes
were retrieved. Twelve were fertilised, and eight day-3 embryos
were available for embryo biopsy. One embryo was found to be
balanced for the FISH signals and heterozygous for
alpha-thalassaemia SEA deletion. That embryo developed to a
good-quality blastocyst of grade 5BB and was transferred. She was
pregnant but refused prenatal invasive testing in the second
trimester because of the risk of miscarriage. She delivered a baby
boy in October 2012 and the cord blood analysis confirmed the
diagnosis of alpha-thalassaemia-1 carrier status with a normal
karyotype 46,XY.
Pre-implantation genetic diagnosis process
Optimisation process (direct mutation detection
and linkage analysis)
The Gap-PCR approach was used to amplify
the alpha-SEA type deletion junction directly (Fig
1). Briefly, the normal allele was amplified by primers
Zdel-1 and Zdel-2 (317 bp), but not by Zdel-1 and Xdel-3, because
they were too far apart. In the alpha SEA deletion, the binding
site for Zdel-2 was deleted, and that for Zdel-1 and Xdel-3 were
brought into close proximity producing a PCR product of 280 bp.
Linkage analysis was performed with fluorescent-labelled
intragenic informative markers (16pTEL05 and 16pTEL06) and linked
short tandem repeats (STR) markers within 2Mb flanking the
alpha-globin loci (D16S521 and D16S3395). The relative positions
of primers and markers around the alpha SEA deletion are shown in
Figure
1. Single cell protocols, using multiple displacement
amplification (MDA) or SurePlex DNA amplification, have been
validated using single lymphocytes from the couple.
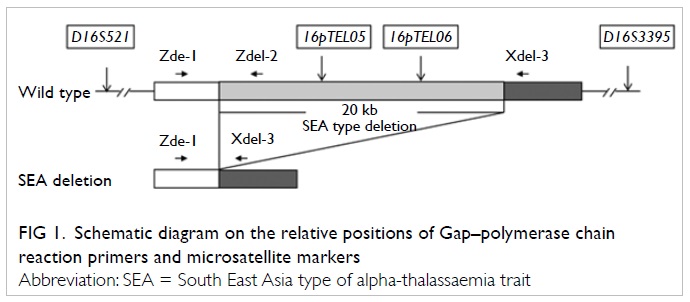
Figure 1. Schematic diagram on the relative positions of Gap–polymerase chain reaction primers and microsatellite markers
Embryo biopsy and pre-implantation genetic
diagnosis
Two blastomeres were biopsied from each of
the good-quality day-3 embryos. One blastomere underwent whole
genome amplification (WGA) and PCR for PGD on the
alpha-thalassaemia loci. The second blastomere was lysed for
translocation detection by FISH.
In the first PGD cycle, WGA was performed
by the MDA method according to the protocol previously published.1 In the second cycle,
SurePlex DNA amplification (BlueGnome) was adopted for WGA. One µL
of WGA product was used for PCR in a final volume of 25 µL
containing 1X PCR buffer with MgSO4, 0.2 mM dNTPs, and
1U FastStart Taq DNA polymerase (Roche). 0.5 µL of PCR product was
separated by an ABI 3500 genetic analyser with a GeneScan
500ROX-size standard (Applied Biosystems) and analysed by
GeneMapper (v4.1; Applied Biosystems).
The second blastomere underwent FISH with
Vysis probes Tel 2p (green), CEP10 (aqua), and Tel 10q (orange).
The signals were interpreted independently by two scientists.
Pre-implantation genetic diagnosis results
In the first PGD cycle, Gap-PCR and
intragenic informative markers 16pTEL05 and 16pTEL06 were used for
PGD. All seven biopsied blastomeres resulted in a conclusive
diagnosis. In the second cycle, the PGD protocol was modified in a
few ways. Firstly, WGA was performed using the SurePlex DNA
amplification system. Secondly, Gap-PCR primers Zdel-1 and Zdel-2
were omitted, since they were poorly amplified in SurePlex WGA
DNA. Finally, two additional linked STR markers (D16S521 and
D16S3395) were used to improve the diagnosis rate. Linkages of
these additional markers with the SEA deletion locus were
established with the leftover WGA DNA from embryos obtained in the
first cycle. All embryos that underwent PGD showed conclusive
results.
Validation of one-blastomere protocol for
double factor pre-implantation genetic diagnosis
The leftover WGA DNA in the second cycle of
PGD was used for array comparative genomic hybridisation (aCGH,
24Sure+, BlueGnome) for the detection of translocation. All
samples showed conclusive result, which was consistent with those
after FISH (Fig 2).
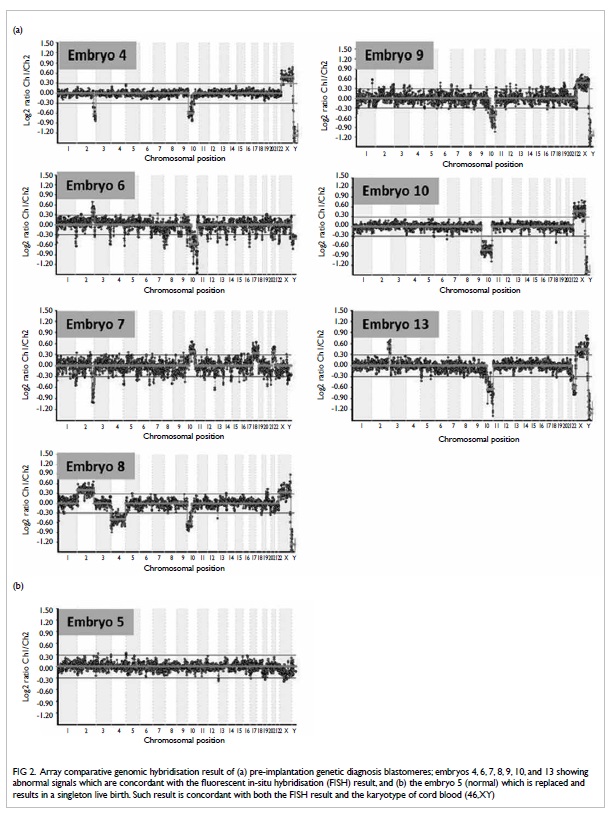
Figure 2. Array comparative genomic hybridisation result of (a) pre-implantation genetic diagnosis blastomeres; embryos 4, 6, 7, 8, 9, 10, and 13 showing abnormal signals which are concordant with the fluorescent in-situ hybridisation (FISH) result, and (b) the embryo 5 (normal) which is replaced and results in a singleton live birth. Such result is concordant with both the FISH result and the karyotype of cord blood (46,XY)
Discussion
Our unit has offered PGD treatment for
monogenetic diseases for more than 10 years, starting in 2000 for
alpha-thalassaemia. The FISH technique was then developed for
translocation carriers and pre-implantation genetic aneuploidy
screening. In this case report, the couple described was the first
to request PGD for both reciprocal translocation and alpha
thalassaemia. The couple firstly attended our unit for PGD in 2009
and at that time the FISH technique was still routinely used for
translocation. We decided to have two blastomeres biopsied, and
undertook PCR on one (for alpha-thalassaemia) and FISH on the
other (for reciprocal translocation). It is well-known that
two-blastomere biopsy is more detrimental than one-blastomere
biopsy on the implantation and pregnancy rate after embryo
transfer.2 Since 2008,
there was emerging evidence regarding the use of array CGH in both
translocation carriers and preimplantation aneuploidy screening.3 4 5 Using aCGH, it could
obtain information on all 24 chromosomes to detect aneuploidy,
which is common in early human embryos, other than in translocated
genetic material.4 Recourse
to WGA in aCGH allowed us to use a single blastomere for both
diagnoses as the amplified products could also be used for PCR. We
switched to using WGA with SurePlex. However, before we acquired
the technique of aCGH for PGD of translocation in 2012, the couple
requested the second treatment cycle because of advancing maternal
age. Therefore FISH was used again in the second PGD cycle for
translocation, as in the first cycle.
Later, we used the leftover WGA DNA from
the second PGD cycle for translocation and aneuploidy detection,
using aCGH 2 weeks after the PGD treatment cycles. All embryos
with abnormal FISH signals showed abnormal aCGH results (Fig
2a). The normal embryo showed a normal signal with no
aneuploidy detected after aCGH, which was performed immediately
after the delivery of the baby boy (Fig 2b). Karyotyping on cord blood of the
baby confirmed our PGD and aCGH results.
So far, there have been three case reports
from the same group of investigators on the use of double-factor
PGD.6 7 8 All of
them involved couples at risk for one genetic disease only (cystic
fibrosis, Von Hippel-Lindau syndrome, Lynch syndrome), but
aneuploidy screening was performed to improve the implantation and
pregnancy rates in those of advanced maternal age. Our patient was
at risk for two genetic diseases, namely alpha-thalassaemia and
reciprocal translocation. In the aforementioned case reports too,
two cells were removed for PGD (either one polar body and one
blastomere, or two blastomeres). Although we also had two
blastomeres biopsied in the treatment cycle of our couple, we have
validated a protocol with which double-factor PGD can be performed
with one-blastomere biopsy. With the use of aCGH, it becomes
feasible and practicable to use one blastomere for both the
monogenetic disease diagnosis and aCGH for either translocation
carriers or aneuploidy screening in at-risk couples. The
turnaround time of our protocol was approximately 2 days,
rendering the fresh cycle day-5 blastocyst feasible for transfer.
Conclusion
We report the first live birth after
double-factor PGD for alpha-thalassaemia and reciprocal
translocation. We have also validated a protocol for double-factor
PGD, in which WGA DNA obtained from a single blastomere can be
used for PCR-based PGD and aCGH.
References
1. Chow JF, Yeung WS, Lau EY, et
al. Singleton birth after preimplantation genetic diagnosis for
Huntington disease using whole genome amplification. Fertil Steril
2009;92:828.e7-10.
2. De Vos A, Staessen C, De Rycke
M, et al. Impact of cleavage-stage embryo biopsy in view of PGD on
human blastocyst implantation: a prospective cohort of single
embryo transfers. Hum Reprod 2009;24:2988-96. CrossRef
3. Wells D, Alfarawati S, Fragouli
E. Use of comprehensive chromosomal screening for embryo
assessment: microarrays and CGH. Mol Hum Reprod 2008;14:703-10. CrossRef
4. Fiorentino F, Spizzichino L,
Bono S, et al. PGD for reciprocal and Robertsonian translocations
using array comparative genomic hybridization. Hum Reprod
2011;26:1925-35. CrossRef
5. Forman EJ, Tao X, Ferry KM,
Taylor D, Treff NR, Scott RT Jr. Single embryo transfer with
comprehensive chromosome screening results in improved ongoing
pregnancy rates and decreased miscarriage rates. Hum Reprod
2012;27:1217-22. CrossRef
6. Obradors A, Fernández E,
Oliver-Bonet M, et al. Birth of a healthy boy after a double
factor PGD in a couple carrying a genetic disease and at risk for
aneuploidy: case report. Hum Reprod 2008;23:1949-56. CrossRef
7. Obradors A, Fernández E, Rius M,
et al. Outcome of twin babies free of Von Hippel-Lindau disease
after a double-factor preimplantation genetic diagnosis:
monogenetic mutation analysis and comprehensive aneuploidy
screening. Ferti Steril 2009;91:933.e1-7.
8. Daina G, Ramos L, Obradors A, et
al. First successful double-factor PGD for Lynch syndrome:
monogenic analysis and comprehensive aneuploidy screening. Clin
Genet 2012;84:70-3. CrossRef

